
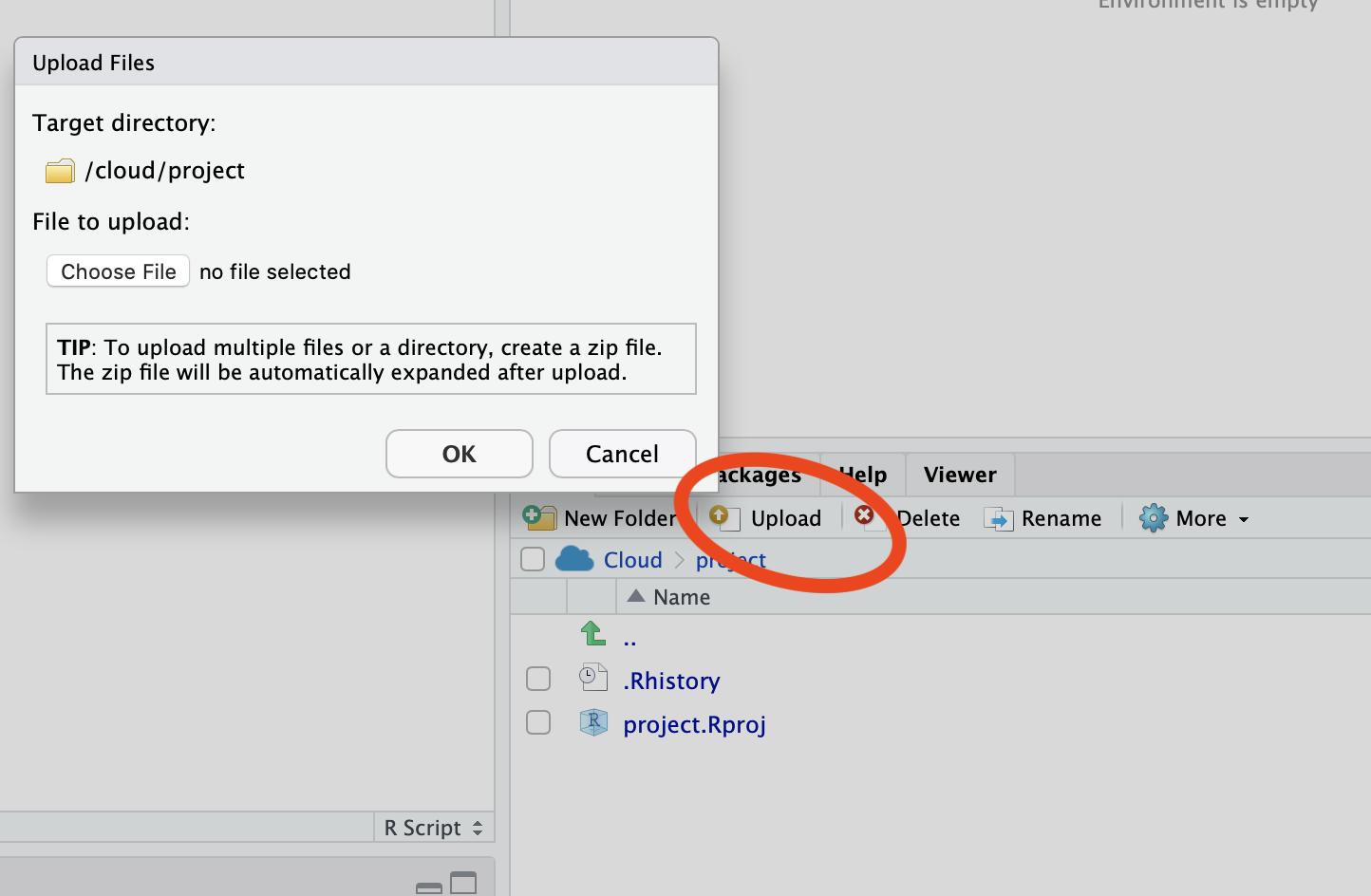
Then, for manipulating these files, you should refer to their tutorial (it's beyond my skills and the scope of your question).For all options above, you may need to install additional packages. Matrix H5I_DATASET 6667 x 24421 H5T_FLOAT

Name obj_type dataset.dims dataset.type_class Lfile = connect(filename = "SCG71.loom", mode = "r+")Īttributes: creation_date, chunks, velocyto._version_, velocyto.logic, version "SCG71:AAACAAACAGCAGAAC" "SCG71:AAACAAACTTTGGGAG"Īfter that, I don't know what to do with that but I think your issue of importing a datafile is because you don't have the right package to open it.Īlternatively, you can also install the package loomR and then enter: library(loomR) "SCG71:AAACGATCCTACTTGT" "SCG71:AAACACTACGTAGAAG" "SCG71:AAACACGGCATCGCAG" You can then load some data > scle$CellID Metadata(4): chunks creation_date velocyto._version_ velocyto.logicĪssays(4): matrix ambiguous spliced unspliced On my computer, once I get it install, I was able to load your SCG71.loom file using: library(LoomExperiment)
On bioconductor, you can also find LoomExperiment. $ ambiguous:Formal class 'dgCMatrix' with 6 slots $ unspliced:Formal class 'dgCMatrix' with 6 slots

$ : chr "SCG71:AAACGATCCTACTTGT" "SCG71:AAACACTACGTAGAAG" "SCG71:AAACACGGCATCGCAG" "SCG71:AAACAAACAGCAGAAC". $ spliced :Formal class 'dgCMatrix' with 6 slots loom file but on the velocyto.R tutorial, it is specified to do the following: library(velocyto.R) I'm not very familiar with this kind of of. loom file that can be open using the velocyto.R package. The file that you are trying to import is not a velocyto file but a. I'm not sure that your issues is from RStudio cloud but more about how you are trying to open the file.


 0 kommentar(er)
0 kommentar(er)
